Abstract
Background
While treatment with the hypomethylating agents (HMAs) azacitidine (AZA) and decitabine (DAC) improves cytopenias and prolongs survival in MDS patients (pts), response is not guaranteed. Identification of non-responders could prevent prolonged exposure to ineffective therapy, avoid toxicities and decrease unnecessary costs. Some genomic abnormalities may predict response to HMAs in small subset of pts, though this approach doesn't take into account the genomic heterogeneity of the disease and the association of these mutations with others.
Aim
We developed an unbiased framework to study the association of several mutations in predicting response to HMAs, analogous to Netflix or Amazon's recommender system in which customers who bought products A and B is likely to buy C: pts who have a mutation in gene A, and B are likely to respond or not respond to HMAs.
Methods
We screened a cohort of 433 pts with MDS (per 2008 WHO criteria) who received HMAs (230 at our institution [training cohort], and 203 at multiple other academic institutions [validation cohort]) for the presence of common myeloid mutations in 29 genes that are obtained by NGS prior to start HMA. Responses were assessed per IWG 2006 criteria. The association between mutations and response was evaluated by the Apriori market basket analysis algorithm. Association rule is a machine learning method that can discover hidden relationships between variables in a dataset. Rules with highest confidence (confidence that the association exist) and highest lift (how strong is the association) were chosen.
Results
Among 433 pts, 193 (45%) received AZA, 176 (40%) DAC, and 64 (15%) received HMA +/- combination. The median age was 70 years (range, 31-100) and 28% were female. A total of 125 (29%) had very low/low risk, 100 (23%) intermediate, 113 (26%) high, and 95 (22%) very high risk category per IPSS-R. Responses included: 95 (22%) complete remission (CR), 14 (3%) marrow CR, 16 (4%) partial remission (PR), and 59 (14%) hematologic improvement (HI). For the purpose of this analysis, pts with CR/PR/HI were considered as responders. The most commonly mutated genes were: ASXL1 (31%), TET2 (22%), SRSF2 (17%), RUNX1 (15%), DNMT3A (14%), SF3B1 (13%), and TP53 (12%). The median number of mutations per sample was 3 (range, 0-9), and 182 pts (42%) had > 3 mutations per sample. In univariate analyses, no single mutation was more prevalent in responders compared to non-responders except NF1 (more common in non-responders, p= .04). TP53 mutations did not predict response or resistance to HMA. A logistic regression multivariate analysis that included clinical and mutational data did not produce a reliable and reproducible model for response. Further, the number of mutations per sample did not predict response (<3 mutations vs. >3 mutations, p= .09). Co-mutated genes such as TET2+ASXL1, TET2+SRSF2, and others did not predict response/resistance to HMA.
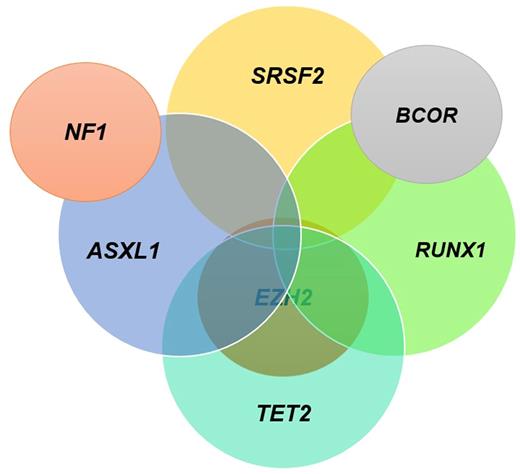
Association rules identified the following genomic combinations as being highly associated with no response: (ASXL1, NF1), (ASXL1, EZH2, TET2); (ASXL1, EZH2, RUNX1), (EZH2, SRSF2, TET2); (ASXL1, EZH2, SRSF2); (ASXL1, RUNX1, SRSF2); (ASXL1, TET2, SRSF2); (ASXL1, BCOR, RUNX1); and (SRSF2, RUNX1, BCOR) while the combination of (TET2, RUNX1, SRSF2) predicted response, Figure 1. These molecular signatures are present in 31% of pts with >/= 3 mutations/sample of the total pt population. They predicted response among 30% of pts with lower-risk disease by IPSS and 30% of pts with higher-risk disease with >/= 3 mutations/sample.
When applying these rules to the validation cohort, the sensitivity of these genomic biomarkers for no response to HMA was 1, specificity for response to HMA was 1, and accuracy was .85.
Conclusion
Genomic biomarkers can identify about a third of pts with MDS who will not respond to HMAs, with high sensitivity. Although these abnormalities are only present in a subset of pts, it can be used to tailor treatment options for these pts by offering alternative therapies to pts with lower risk disease and enrolling pts with higher risk disease on clinical trial with novel combination or proceed to transplant without prior HMA treatment. This study highlights the complexity of interpreting genomic data in regards to response to HMA and addresses the importance of machine learning technologies such as the recommender system algorithm in translating genomic data into useful clinical tools.
Sekeres: Celgene: Membership on an entity's Board of Directors or advisory committees. Bejar: Genoptix: Consultancy, Honoraria, Patents & Royalties; Celgene: Consultancy, Honoraria, Other: DSMB, Steering Committee, Research Funding; Otsuka/Astex: Honoraria, Other: Ad-hoc advisory board; Foundation Medicine: Honoraria, Other: Ad-hoc advisory board; AbbVie/Genetech: Honoraria, Other: Ad-hoc advisory board; Modus Outcomes: Consultancy, Honoraria. Komrokji: Novartis: Honoraria, Speakers Bureau; Celgene: Honoraria. Steensma: Onconova: Consultancy; Janssen: Consultancy, Research Funding; Takeda: Consultancy; Incyte: Equity Ownership; H3 Biosciences: Consultancy; Celgene: Consultancy; Pfizer: Consultancy; Amgen: Consultancy, Membership on an entity's Board of Directors or advisory committees; Pfizer: Consultancy, Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees. Roboz: Cellectis: Research Funding; AbbVie, Agios, Amgen, Amphivena, Array Biopharma Inc., Astex, AstraZeneca, Celator, Celgene, Clovis Oncology, CTI BioPharma, Genoptix, Immune Pharmaceuticals, Janssen Pharmaceuticals, Juno, MedImmune, MEI Pharma, Novartis, Onconova, Pfizer, Roche Pharmace: Consultancy. Ebert: H3 Biomedicine: Consultancy; Genoptix: Patents & Royalties; GRAIL: Consultancy; Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal